BACKGROUND:
Mitochondria are important in the pathology of sickle cell disease (SCD). They are abnormally retained in sickle red blood cells (RBCs) and the likely source of cell-free mitochondrial DNA (mtDNA), an erythrocytic-DAMP that triggered formation of neutrophil extracellular traps (Tumburu et al, 2022). Acquired mutations in the mitochondrial genome, referred to mtDNA heteroplasmy, are emerging markers of aging and inflammation. Using whole genome sequence data from peripheral blood DNA, we previously established that patients with SCD not only have increased mtDNA heteroplasmy, but the burden also varied with the sickle genotype [Ahmad et. al, 2021]. The burden of mtDNA heteroplasmy is known to vary across different organs and tissues. Here, we utilized the Townes mouse model of SCD to explore differential mtDNA heteroplasmy burden across the different tissues and sickle genotypes in SCD.
METHODS:
Genomic DNA was extracted from 9 different organs (heart, brain, muscle, kidney, blood, bone marrow, liver, lungs, spleen) of littermate mice (3 male and 3 female) of three genotypes (HbAA, HbAS, HbSS) in three replica sets (total number of mice = 18, 9 different tissue samples per mouse, total sample number = 162). Mitochondria DNA was enriched from genomic DNA by long range PCR in 2 amplicons of 9kb and 7kb sizes (Misa Hirose et al, 2018). The enriched mtDNA PCR amplicons were mixed together and subjected to library preparation for deep sequencing using Illumina platform. Sequence reads aligned to mtDNA were mapped and called for variants using LoFreq variant calling tool.
RESULTS:
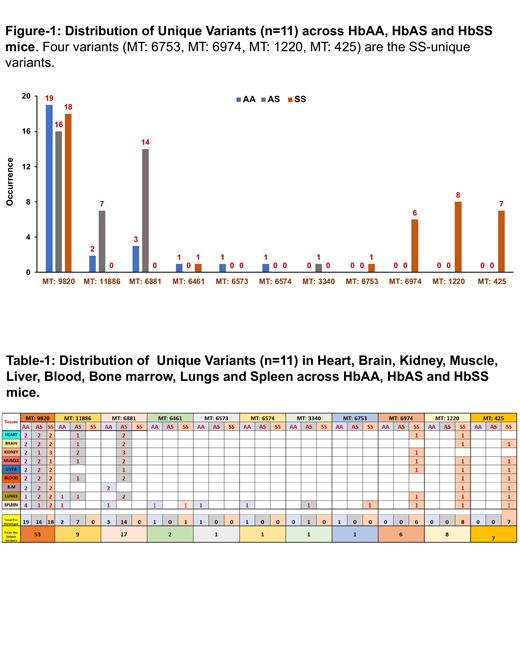
Of the 162 samples, 1 sample (HbAA, lung) was excluded from analysis due to low coverage. Using LoFreq variant caller tool (VAF cut-off 4% & Coverage Filter 3500X), and after removing the repeating variants, 11 unique variants spanning 16kb mouse mitochondrial genome, were found in 106 of the 161 (65.8%) samples. The variants were differentially distributed across the tissues and genotypes with the variant at mito-position 9820 (MT: 9820) being most frequent, present in 19, 16 and 18 samples in AA, AS and SS mice, respectively (Fig. 1). Two variants at MT: 11866 and MT: 6881 were shared between AA and AS mice. Four variants were unique to SS mice: MT: 6753 in COX1 gene causing Phe476-Leu substitution, MT: 6794 in intergenic region upstream to COX2 gene, MT: 1220 in intergenic region upstream to ND1 gene, and MT425 in regulatory/D-Loop region (Table -1). The SS-unique variants were present in all tissues except for Cox1 Phe476-Leu (MT: 6753), a single variant that was found only in the spleen. Overall, HbSS had the highest concentration of variants distributed among 41/54 (75.9%) followed by AS 38 /54 (70.3 %) and AA 27/53 (50.9%). Female mice had higher mtDNA heteroplasmy burden compared to male mice in all genotypes. Across all genotypes and tissues, spleen had the highest number of variants (16.03%) and liver, the lowest (9.4%).
CONCLUSION:
In keeping with data in patients with SCD, HbSS mice had the highest mtDNA heteroplasmic burden followed by HbAS and HbAA genotypes. Four mito-variants were unique to HbSS. We also found differentially higher mtDNA heteroplasmic burden in splenic tissues and speculate that this could arise from metabolic stress conditions in the spleen.
Disclosures
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal